Other Projects
Table of contents:
- EQUIP Project
- software populations approach to ubicomp systems design
- Bayesian analysis of Raman spectroscopy
- Bayesian methods to detect dye-labelled DNA oligonucleotides in multiplexed Raman spectra
- Preferential attachment of specific fluorescent dyes and dye labelled DNA sequences in a SERS multiplex
- Bayesian modelling and quantification of Raman spectroscopy
- Unbiased local solutions of partial differential equations via the Feynman-Kac Identities
- Uncertainty in models of alignment
EQUIP Project
software populations approach to ubicomp systems design
Bayesian analysis of Raman spectroscopy
This research project aims to develop new methods for modelling and quantification of surface-enhanced Raman spectroscopy (SERS). Spectra are multivariate observations of the interaction between light and matter. Raman spectroscopy can be used to identify molecules such as DNA by the characteristic scattering of light from a laser. It is sensitive at very low concentrations and can accurately quantify the amount of a given molecule in a sample. Metallic nanoparticles are used in SERS to enhance the Raman signal. The presence of a large, nonuniform background presents a major challenge to analysis of these spectra.
This project is funded by an EPSRC programme grant (ref: EP/L014165/1)
https://www.slideshare.net/azeari/bayesian-modelling-and-computation-for-raman-spectroscopy
Collaborators on this project;
- Duncan Graham (University of Strathclyde) - Principal Investigator
- Mark Girolami (Imperial & Turing)
- Karen Faulds (Strathclyde)
- Pasquale Maffia (Glasgow)
- Kirsten Gracie (Strathclyde)
- Matt Moores (Warwick)
- Jake Carson (Imperial)
- Steven Asiala (Strathclyde)
- Jonathan Noonan (Glasgow)
Bayesian methods to detect dye-labelled DNA oligonucleotides in multiplexed Raman spectra
Inferential methodologies are required which can deconvolve the observed mixture and infer the composition of distinct DNA sequences in the overall composite. Inferring the component spectra is posed as a model selection problem for a bilinear statistical model, and the Markov chain Monte Carlo inferential methodology required is developed. In particular, a Gibbs sampler and reversible jump Markov chain Monte Carlo (RJ-MCMC) methods are presented along with techniques based on estimation of the marginal likelihood.
- Reference: Zhong, M.; Girolami, M.; Faulds, K. & Graham, D. (2011) Bayesian methods to detect dye-labelled DNA oligonucleotides in multiplexed Raman spectra J. R. Stat. Soc. Ser. C, 60, 187-206. https://doi.org/10.1111/j.1467-9876.2010.00744.x
Preferential attachment of specific fluorescent dyes and dye labelled DNA sequences in a SERS multiplex
Here, the interaction of single stranded DNA labeled with either fluorescein (FAM) or tetramethylrhodamine (TAMRA) with a metal surface, using spermine induced aggregated silver nanoparticles as the SERS substrate, is investigated by analyzing the labels separately and in mixtures. When the two dyes are premixed prior to the addition of nanoparticles, TAMRA exerts a strong masking effect over FAM due to a stronger affinity for the metal surface. By using bootstrap estimation of changes in SERS peak intensity, a greater insight has been achieved into the surface affinity of the two dyes as well as how they interact with each other. It has been shown that the order of addition of the analytes is important and that specific dye related interactions occur, which could greatly affect the observed SERS spectra.
Reference: Gracie, K.; Moores, M.; Smith, W. E.; Harding, K.; Girolami, M.; Graham, D. & Faulds, K. (2016) Preferential attachment of specific fluorescent dyes and dye labelled DNA sequences in a SERS multiplex Anal. Chem., 88, 1147-1153. https://doi.org/10.1021/acs.analchem.5b02776
Bayesian modelling and quantification of Raman spectroscopy
We introduce a sequential Monte Carlo (SMC) algorithm to separate the observed spectrum into a series of peaks plus a smoothly-varying baseline, corrupted by additive white noise. The peaks are modelled as Lorentzian, Gaussian, or pseudo-Voigt functions, while the baseline is estimated using a penalised cubic spline. This latent continuous representation accounts for differences in resolution between measurements. The posterior distribution can be incrementally updated as more data becomes available, resulting in a scalable algorithm that is robust to local maxima.
Preprint: Moores, M.; Gracie, K.; Carson, J.; Faulds, K.; Graham, D. & Girolami, M. Bayesian modelling and quantification of Raman spectroscopy https://arxiv.org/abs/1604.07299
Unbiased local solutions of partial differential equations via the Feynman-Kac Identities
The Feynman-Kac formulae (FKF) express local solutions of partial differential equations (PDEs) as expectations with respect to some complementary stochastic differential equation (SDE). In this paper we utilize recent developments in two areas to demonstrate that it is now possible to obtain unbiased solutions for a wide range of PDE models via the FKF. The first is the development of algorithms that simulate diffusion paths exactly (without discretization error), and so make it possible to obtain Monte Carlo estimates of the FKF directly. The second is the development of debiasing methods for SDEs, enabling the construction of unbiased estimates from a sequence of biased estimates.
Preprint: Carson, J.; Pollock, M. & Girolami, M. Unbiased local solutions of partial differential equations via the Feynman-Kac Identities https://arxiv.org/abs/1603.04196
Uncertainty in models of alignment
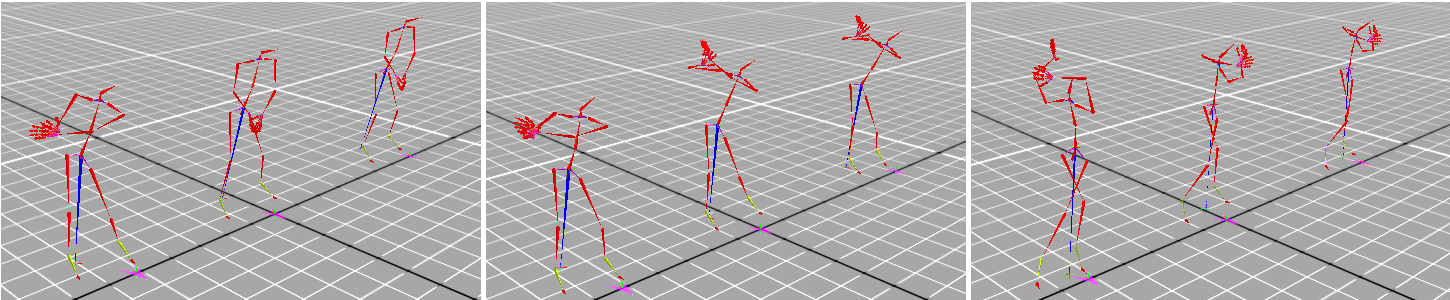
Temporal alignment of sequences is the task of removing the differences between the observed time series arising from the differences in their relative timing. It is a common preprocessing step in time series modelling, usually performed in isolation from the data analysis and modelling. We propose casting alignment learning in a framework where both the alignment and the data is modelled simultaneously. Combined with a probabilistic alignment objective, such an approach allows us to align sequences into multiple, a-priori unknown groups in an unsupervised manner. Furthermore, the use of Bayesian nonparametrics offers the benefits of principled modelling of the noisy observed sequences, explicit priors that encode our beliefs about the constituent parts of the model and the generative process of the data, and an ability to adapt to the complexity of the data.
Another feature of the probabilistic formulation that is lacking in the traditional temporal alignment models is an explicit quantification of the different kinds of uncertainties arising in the alignment problem. These include the uncertainties related to the noisy observations and the fact that the observed data may be explained in multiple different ways, all of which are plausible under our prior assumptions. While formulating various parts of the model, we encounter and discuss some of the challenges of Bayesian modelling, most notably the need for approximate inference. We argue that variational distributions which include correlations between the hierarchical components of the model are necessary to take advantage of the potential of the model to discover the compositional structure in the data and to capture the uncertainty arising from it.
Collaborators on this project:
- Neill D.F. Campbell (University of Bath)
- Carl Henrik Ek (University of Cambridge)
- Ivan Ustyuzhaninov (University of Tuebingen)
- Markus Kaiser (TU Munich)
- Erik Bodin (University of Bristol)